Exercise 1 - Nuclei segmentation
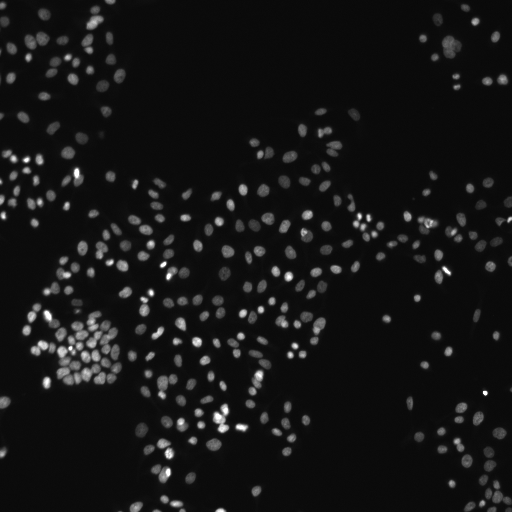
Human HT29 colon cancer cell nuclei, image from Broad Bioimage Benchmark Collection
Global segmentation
Open above image by dragging it into the Fiji window and run Threshold command:
Image > Adjust > Threshold...Choose different thresholding methods (such as Default, Huang, Otsu etc.) from the drop down list and check how well do they segment nuclei in the image.
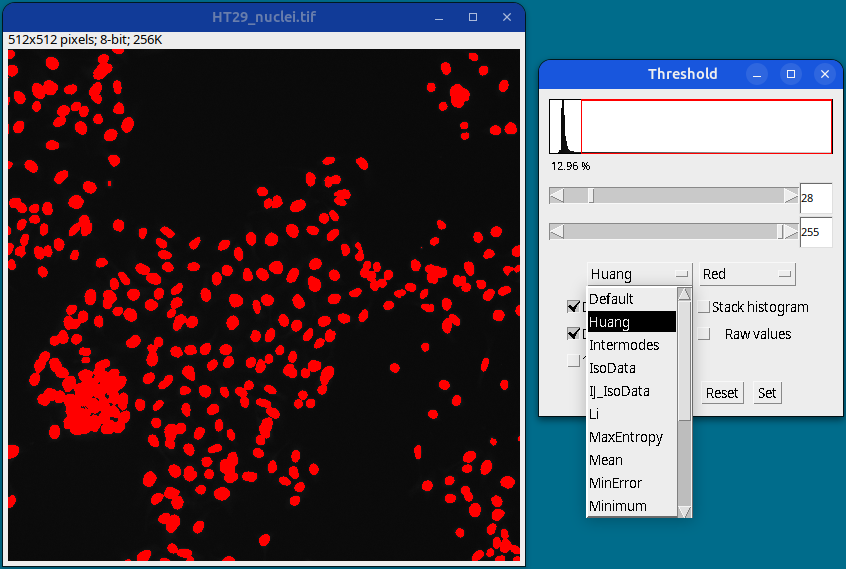
Once satisfied with a particular method or by manually selecting the lower and upper threshold values (using sliders), click on the Apply button to generate a thresholded image.
All thresholding methods (such as Default, Huang, Otsu etc.) can be tested at once by using Image > Adjust > Auto Threshold
Local segmentation
Select your original image and run the command:
Image > Adjust > Auto Local Threshold
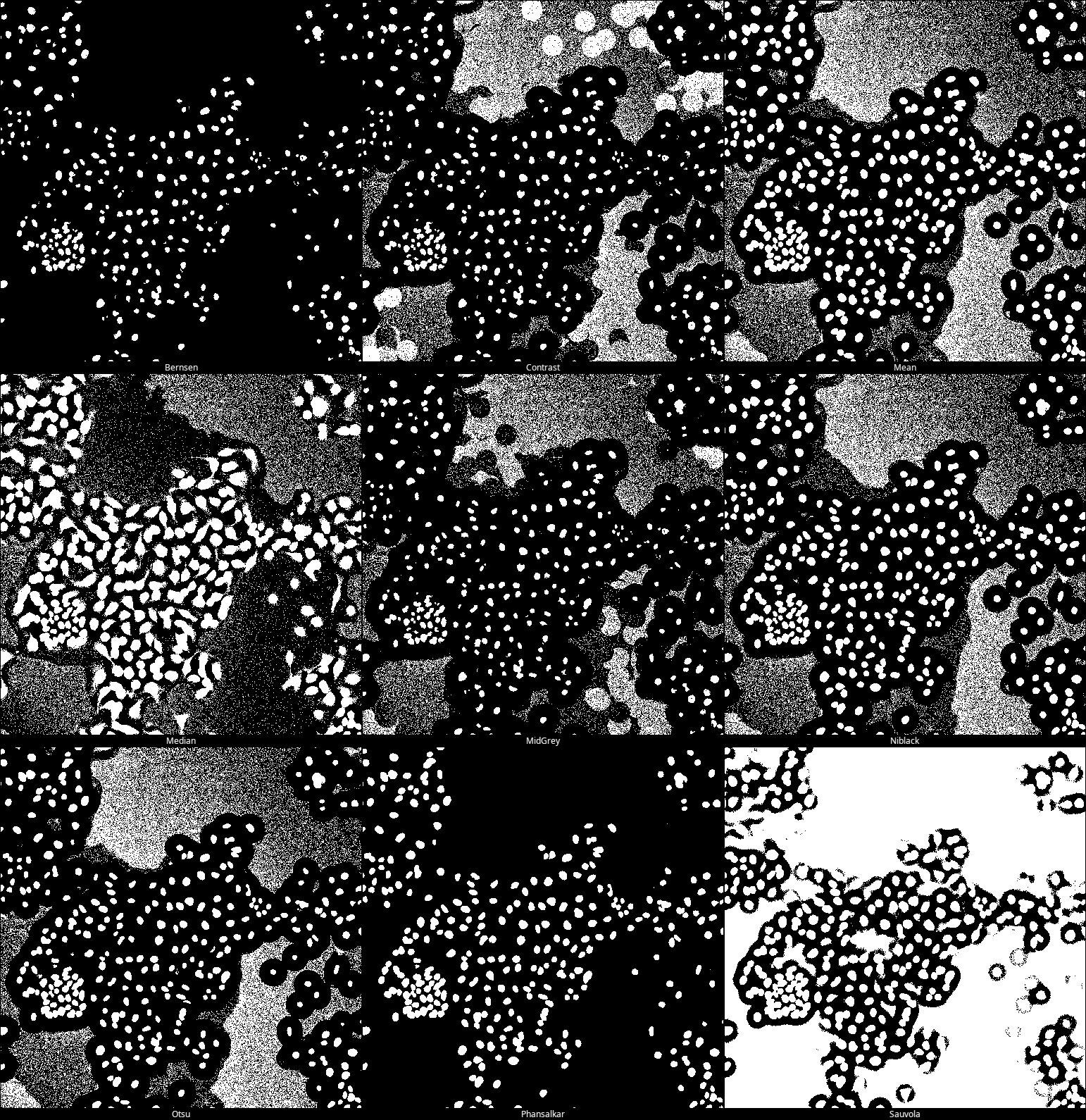
Run with “Try all” method to check which one gives the best result.
For this image, the best segmentation is achieved with the Phansalkar method (bottom row, middle image).
Deep Learning based segmentation using StarDist
Select your original image and run the command:
Plugins › StarDist › StarDist 2DIn the follow up StarDist menu, choose Model:
Versatile (fluorescent nuclei),
and click on theSet optimized postprocessing thresholdsbutton at the bottom.
Keep other settings as deafult. Click OK.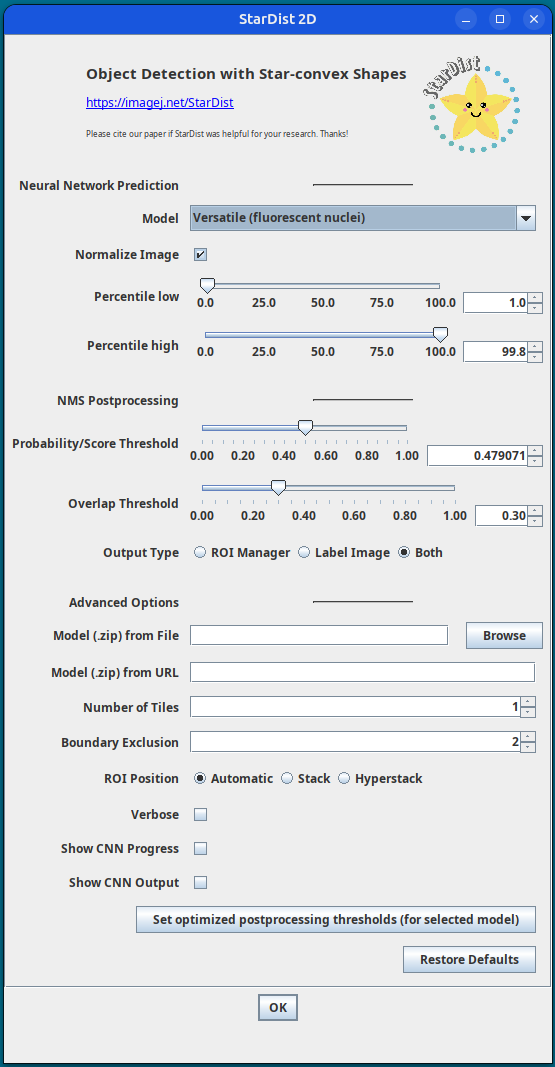
StarDist settings A segmentation label image will be generated with the nuclei ROIs added to the ROI Manager.
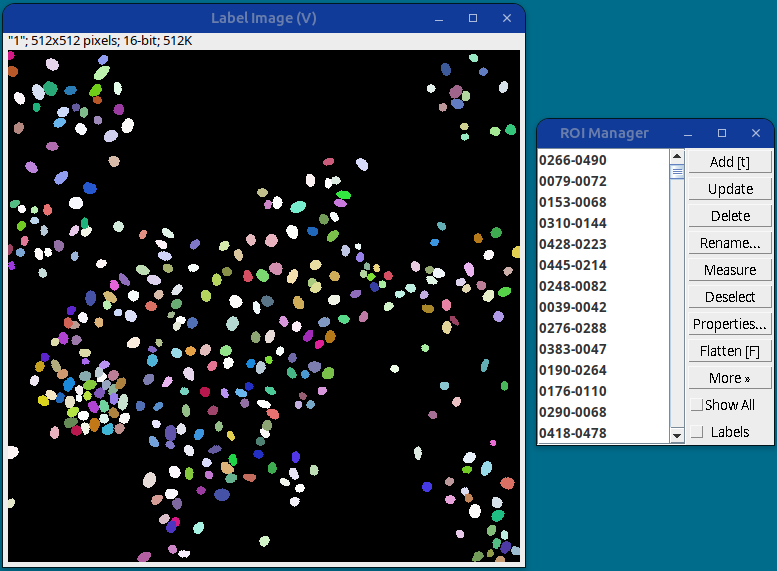
StarDist segmentation results This gives the best nuclei segmentation among all the methods tested so far.